Cell Line Development – Data Handling
Summary
- Cell line development requires sample data be:
- Collated from different analyzers and formats
- Tracked between different plates and wells
- Maintained and accessed over weeks or months
- Biomek and DART software can handle this process behind the scenes of an automated process
- Data is reliably stored for traceability and easily accessed without the need for IT intervention
When automating a workflow, one often only thinks about the liquid handling aspects required to handle higher throughputs or reduce the time at the bench. However, the data that is generated from any workflow is what is truly valuable and yet still somehow an afterthought when it comes to automation.
Cell line development is one of many workflows that has significant data challenges. Like any screening process, the sample throughput provides a challenge from the sheer amount of data. This becomes even more challenging when multiple analytical instruments are used to generate different types of data, often in different formats. While all screens have some aspect of hit identification, the hit picking workflows require that the data must be tracked with these hits. Not only does this involve moving data across different plates and wells, but the overall process can extend for months. The data must be traced throughout this process to ensure that any cell lines that move into manufacturing have the requisite historical data (e.g. proof of monoclonality) and that all data is easily accessed for reporting needs.
Automation can help ensure accurate data tracking throughout a lengthy workflow, such as the monthslong multi-step cell line development workflow (Figure 1). While plate barcodes can be scanned on a variety of analyzers, manual workflows still resort to SOPs and training as a way to deal with the data challenge. However, copying and reformatting data in Microsoft Excel is inherently an errorprone process, particularly when trying to correlate data from different analyzers. For example, the CloneSelect Imager, which we use for monoclonality and growth determination, reports results by rows (A1, A2, A3…Figure 2A). In contrast, the Octet HTX we used to titer IgG production reports results by column (A1, B1, C1…, Figure 2B). If one tries to re-sort the Octet data in Microsoft Excel by well name, the wells still do not align with the CloneSelect data (A1, A10, A11…, Figure 2C). Collating different analyzer data in a single format is just one example of data pitfalls. Here we describe how automating cell line development on a Biomek i7 Workstation (Figure 3) overcame the many data challenges associated with this workflow.
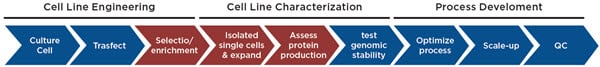
Figure 1. Cell line development process. The data management for the highlighted steps are described here.
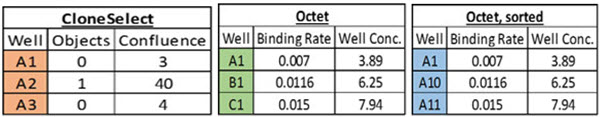
Figure 2. Data correlation. A) Imaging data from the CloneSelect Imager is reported in row-wise fashion. B) IgG titer data from the Octet HTX is reported in column-wise fashion. C) Attempting to “sort” the Octet data by Well still does not match the row-wise list of the CloneSelect for correlation.

Figure 3. The Biomek i7 Workstation, along with various integrated devices, was used to automate a cell line development workflow.
While one would expect any automation system to track and utilize data such as well volumes, storing and utilizing assay data adds another level of complexity. For the cell line development process, meaningful data was first generated when we assayed colony characteristics in semi-solid media on a ClonePix 2 device. During picking, the initial characteristics identified by the ClonePix 2, such as colony size and external fluorescence (a marker of antibody secretion) are linked to a barcoded destination plate and well. This data can be automatically imported into the Biomek software when the barcode is scanned. The data can then move with the samples as Biomek software automatically moves data as part of standard liquid transfer steps (Figure 4A). The data can also be stored in the DART database, and any time the plate barcode is scanned in the future, that corresponding data is automatically retrieved. DART provides the link between methods that are staggered over time (Figure 4B) – a key component in a lengthy workflow such as cell line development.
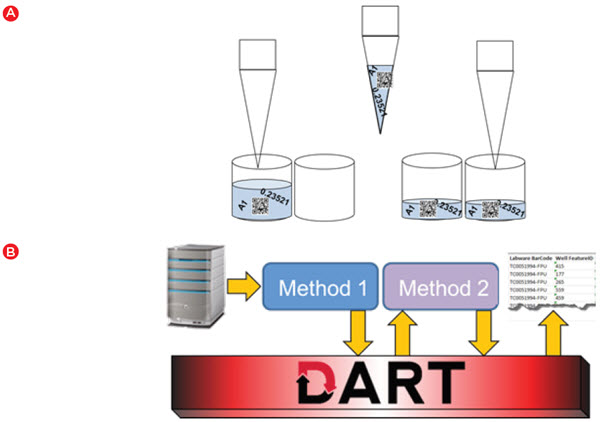
Figure 4. Automated data handling. A) Biomek software automatically transfers data from source to destination when a liquid handling step is executed. B) DART software provides a database that can be used to store data between runs and enables data to be accessed in a later run or for generating reports.
To further illustrate the data challenges of the cell line development workflow, we will describe a single day of the months-long process, and describe how these challenges were overcome through automated data software tools. Three weeks after plating cells by limiting dilution and identifying monoclonal wells, data moves and is used in the following steps (Figure 5):
- The barcoded plates are scanned as the move from the integrated incubator to the Biomek and a “Load Data” step in the method retrieves previous data associated with this plate. These include monoclonality data but also any earlier data deemed relevant such as ClonePix colony size and external fluorescence.
- The plates are scanned for confluence on the CloneSelect Imager and a “Create Data Set” step attaches this well-level data to the barcoded plate.
- Because we only want to assay protein titer from wells with colonies grown from a single cell, we can create a hit picking data set to identify only these wells for sampling. This data set (“MonoGrowth”) can drive the pipetting in a Biomek transfer step. The media from the sampled wells was consolidated and reformatted into 384-well Octet assay plates.
- The 384-well plate was assayed on the Octet and the resulting IgG concentrations were attributed to the assay wells through another “Create Data Set” step.
- Using a “Copy Data Set” step, the Octet assay results were transferred back into the original 96-well plates containing the colonies. This deconvolution step automatically handles the previously described problem of row vs. column lists.
- The new data is used to drive another hit picking transfer step such that the monoclonal wells with IgG expression above a set threshold are transferred to 24-well plates for continued expansion and testing. All of the essential data (monoclonality, confluence, IgG levels, etc.) are automatically moved forward to the new wells so there is no need to trace back to the original 96-wells to find the relevant data.
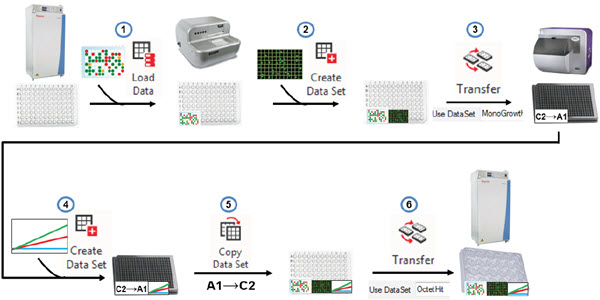
Figure 5. Automated data handling. Biomek and DART software can combine to retrieve previous data, add new assay data to plates, drive reformatting transfer steps, copy assay data back to the source wells, to ensure the proper hits are brought forward.
These steps of linking assay data to sample wells, correlating data from multiple analyzers, using data to drive transfers or hit picking, and bring all data forward for traceability, are repeated throughout the cell line development process. In addition, storing this data for all potential cell lines is essential as many will drop out at later points due to genomic instability or other undesirable attributes and it is impossible to know which will ultimately be the lines selected for cell banking.
Storing and utilizing data for automated runs is only part of the story. You must also be able to easily access the data when needed and the DART database can be accessed through multiple convenient means. For quick visualization in a user-friendly manner, the DART Data Browser (Figure 6A) can highlight method-, labware-, and well-level data in a standard Google Chrome Browser. Not only can this data be viewed remotely, but it can be refreshed to update with live data during runs. For exporting data in a usable form, the DART Report Builder can be used to generate reports in Microsoft Excel, with a simple interface to choose the data to include in a given report. All of this can be done without any assistance from IT resources – a common requirement for some larger data management systems.
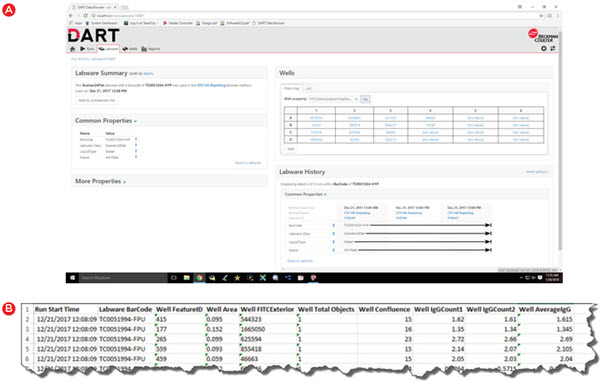
Figure 6. Data viewing and reporting. Data must be easily accessed as needed and DART enables viewing from a standard web browser (A) or selection of data to be included in Microsoft Excel-based reports (B).
The data generated during any experiment is what makes that experiment worthwhile. Any errors in data handling can eliminate this value or perhaps even send a research program in the wrong direction. For a months-long workflow like cell line development, the risks of data loss or errors are even more significant, thus, tools to ensure data integrity are paramount. By making this data integrity process automatic and easily accessible, Biomek and DART software can provide true value to the cell line development process.
Biomek Automated Workstations are not intended or validated for use in the diagnosis of disease or other conditions. Beckman Coulter Life Sciences genomic reagent kits are for research use only.


